CountBio
Mathematical tools for natural sciences
Biostatistics with R
Multiple curves on the same plot
To plot more than one curve on a single plot in R, we proceed as follows. Create the first plot using the
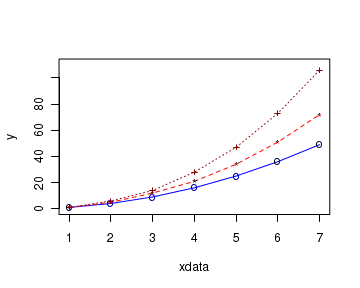
In the R script shown below, we plot the curves of three data sets (x,y1),(x,y2) and (x,y3) on the same plot. The code and the plot are shown here:
# define 3 data sets xdata <- c(1,2,3,4,5,6,7) y1 <- c(1,4,9,16,25,36,49) y2 <- c(1, 5, 12, 21, 34, 51, 72) y3 <- c(1, 6, 14, 28, 47, 73, 106 ) # plot the first curve by calling plot() function # First curve is plotted plot(xdata, y1, type="o", col="blue", pch="o", lty=1, ylim=c(0,110) ) # Add second curve to the same plot by calling points() and lines() # Use symbol '*' for points. points(xdata, y2, col="red", pch="*") lines(xdata, y2, col="red",lty=2) # Add Third curve to the same plot by calling points() and lines() # Use symbol '+' for points. points(xdata, y3, col="dark red",pch="+") lines(xdata, y3, col="dark red", lty=3)
Adding legends to the multiple curves
Legends can be added to describe the different curves in the plot. For this, we call the
x, y -----> X and Y axis locations in the graph coordinates. legend-----> A vector of string consisting of legends, typically one per graph col -----> A vector of colors for 'col' parameter. These colors are same as the ones used in the graph pch -----> A vector of character symbols for 'pch' parameter, same as the ones used as 'pch' parameters in the plots. lty -----> A vector of line types to be given to 'lty' parameter, same as the one used for plotting curved ncol -----> Number of columns for writing the legends. Default is one column. For other parameters, type help() in R prompt. While typing the vectors for legend, col and pch, the order of the plot types have to be maintained.
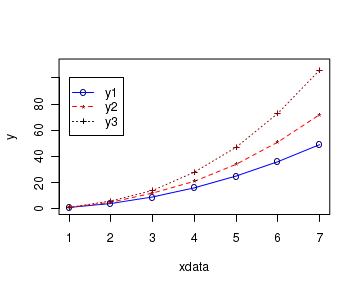
Carefully note the example code below which adds legend to the plot. The resulting figure has three curves with a legend box:
# define 3 data sets xdata <- c(1,2,3,4,5,6,7) y1 <- c(1,4,9,16,25,36,49) y2 <- c(1, 5, 12, 21, 34, 51, 72) y3 <- c(1, 6, 14, 28, 47, 73, 106 ) # plot the first curve by calling plot() function # First curve is plotted plot(xdata, y1, type="o", col="blue", pch="o", lty=1, ylim=c(0,110), ylab="y" ) # Add second curve to the same plot by calling points() and lines() # Use symbol '*' for points. points(xdata, y2, col="red", pch="*") lines(xdata, y2, col="red",lty=2) # Add Third curve to the same plot by calling points() and lines() # Use symbol '+' for points. points(xdata, y3, col="dark red",pch="+") lines(xdata, y3, col="dark red", lty=3) # Adding a legend inside box at the location (2,40) in graph coordinates. # Note that the order of plots are maintained in the vectors of attributes. legend(1,100,legend=c("y1","y2","y3"), col=c("blue","red","black"), pch=c("o","*","+"),lty=c(1,2,3), ncol=1)